Introduction
Overview
Teaching: 10 min
Exercises: 0 minQuestions
Why standardize our taxonomic collections?
What tools should we use to standardize our collections?
Objectives
Understand that there are a lot of catalogues available for standardizing taxonomy
Understand that there are multiple tools available for name alignment
What is Taxonomic Name Alignment?
Aligning taxonomy is a process of assuring the names in our collections are the most current and up to date according to an authority of interest. It involves comparing a list of inputed names to the authority’s database and returning an output of resolved names.
Why is Taxonomic Alignment Important?
Scientific names and the species they represent are hypotheses. As in all fields of science, these hypotheses should always be refined based upon the most current and accurate information. Species in the past may have been identified based upon a particular trait or an authority’s species concept. As we collect more data from fields like systematics and population genetics, we are more capable of refining these designations to represent our most current knowledge. Its therefore important to standardize our collections to the most up to date designations based upon new information.
What Taxonomic Resources can we use to Align Names?

Taxonomic databases that contain name designations are called “catalogues”. Catalogues contain the necessary information for aligning old names with their most current counterparts if they have changed. Due to scope, species concepts, and project aims there are quite a few catalogues to choose from when aligning names. Some catalogues are designed in large scope to encompass a large portion of the tree of life, while others are more focused on particular clades. Depending on the group you are working with, different catalogues may offer more up to date expert designations. It is important to note that catalogues’ name designations are hypotheses. They are not the absolute truth and are subjective to how the catalogue builds their interpretations. Available data & how we study particular organisms can change how catalogues establish what is and is not a species. To address these issues, it is important to consider the following:
- What type of catalogue am I using? Is it a catalogue that specializes in a particular clade/group? Does it aggregate other catalogue’s interpretations?
- What methods does the catalogue use to build its taxonomic backbone? Expert opinion? Phylogenetic trees? A combination of methods?
- How complete is the catalogue? How current are the designations?
How are Catalogues Structured?
There are various formats depending on the catalogue, but in general they all share the feature of linking names by a relationship. Two common relationships we will see are:
- Accepted Name: This relationship shows that the name searched is registered as the most current/valid name according to the catalogue.
- Synonym: This relationship suggests that the name searched has been remapped to an alternative name according to the catalogue.

The Name-Alignment-Tool
Rather than going by hand and correcting your collection’s names manually, there are tools available to align lists of hundreds to thousands of names to catalogue’s standards. The tool we’ll be showcasing today takes advantage github’s interface, allowing a graphical type enviroment that many of us are used to. Below is a diagram providing an overview of how this tool works. We will begin addressing each of these steps one by one, but first a light introduction to Github.
More Info: How Does this tool Work?If you are interested in how the tool actually aligns these names you can visit the repository for Nomer. Nomer is the tool used for aligning names within GloBI.
Next Up: Light Introduction to Github
Key Points
Choosing your catalogue to align your collection’s names to is important!
The name-alignment-tool is an option that requires no programming experience and reduces overhead that scripting languages are prone to
Navigating Github and Creating a Copy of the Tool
Overview
Teaching: 15 min
Exercises: 0 minQuestions
What is Github?
What are the basics of its interface we need to know to use the tool?
Where and how do I access the name-alignment-tool?
Objectives
Understand your github account, what a repo is, and how to navigate a repo
Creating your own copy of the name-alignment-tool
Getting started
To use Github you will need a github account as mentioned in the setup instructions. Please note you do not need Git downloaded onto your computer for this tutorial. We will be working in Github’s graphical interface for the purpose of this lesson.
What is Github?
Simply put github is a hosting service for software development and collaboration. Some of us use it to store versions of our code/analyses and others use it for development of software/applications. Today our aim isn’t to utilize github for these aspects, but to interact with the name-alignment-tool that has been built on github. All you need to interact with the tool is to make a copy of the template repository. That probably sounds complicated at first so lets break it down into a few steps.
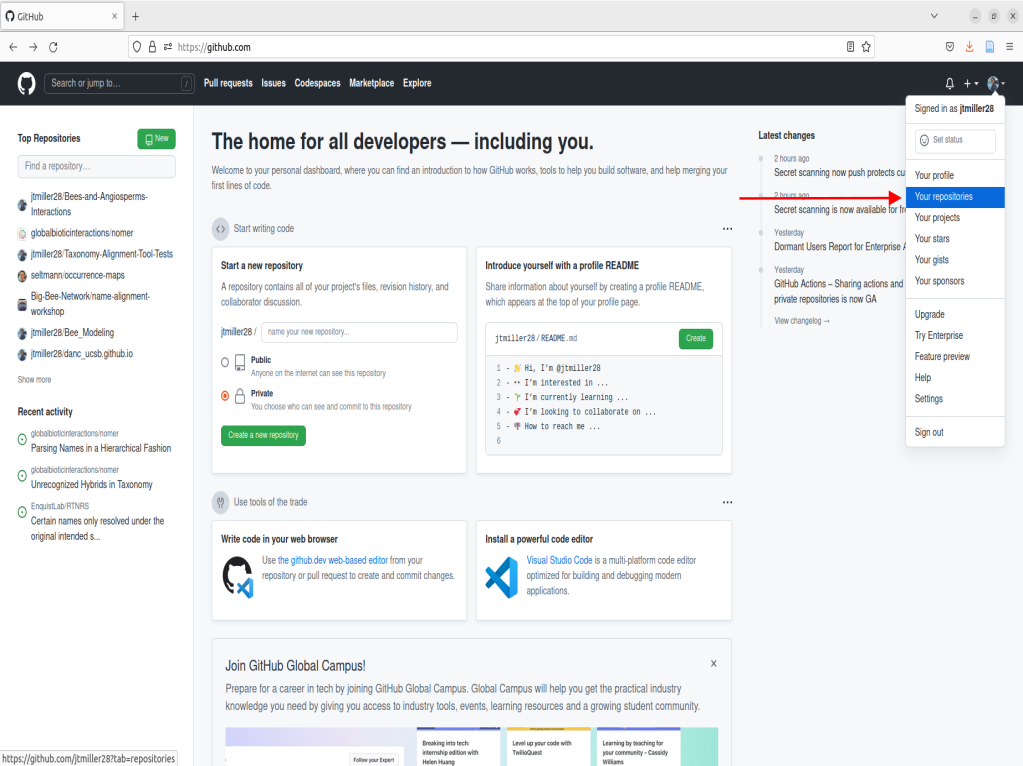
Once you have made you’re account, you will likely be taken to a page showing you’re home activity/github’s recommendations. From there, we can nagivate to the top right corner that displays your profile icon, click it, then navigate to the tab that says “Your repositories”, go ahead and click that as well.
What is a Repository or “Repo”?
Right now you should see the following page after navigating to your repositories:
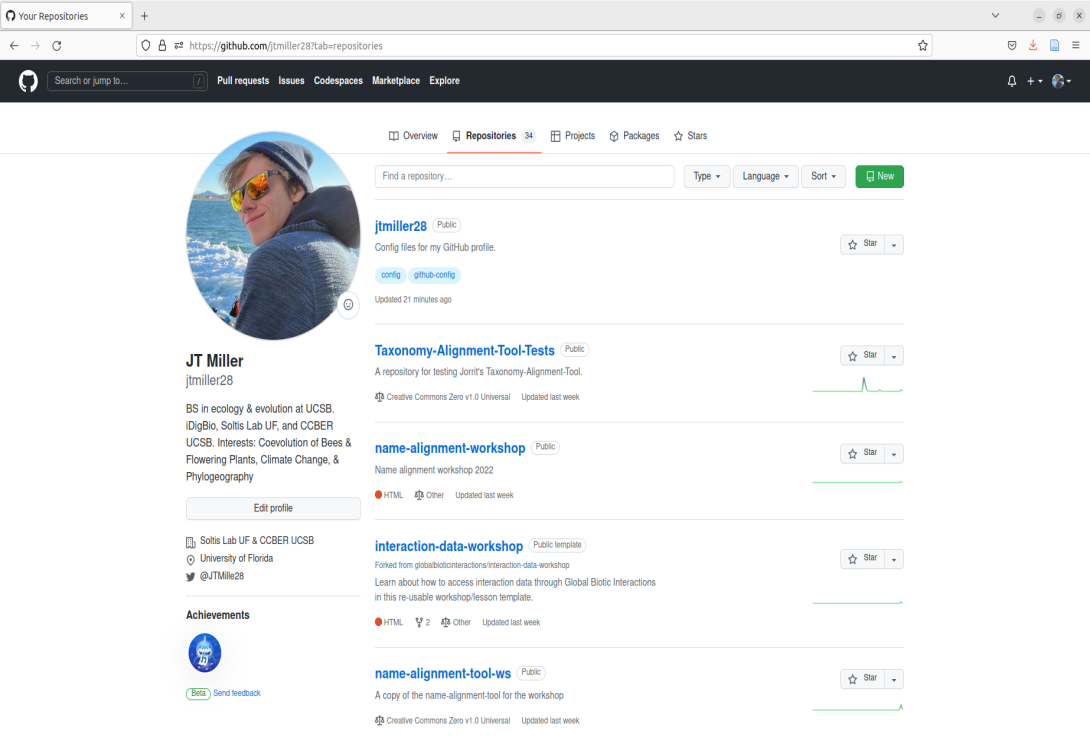
This page contains what are called Repositories on GitHub or for short “Repos”. A repository is a location on GitHub that hosts a collection of files associated with your work. It is like a folder holding all the files for an associated project. The page we are currently on shows your repositories. If you are new to GitHub, you currently will not have any repositories listed on this landing page. Rather than building a repository completely from scratch, we are going to create a copy of one already built. To do this we can navigate to the repo that is the template for the name-alignment-tool.
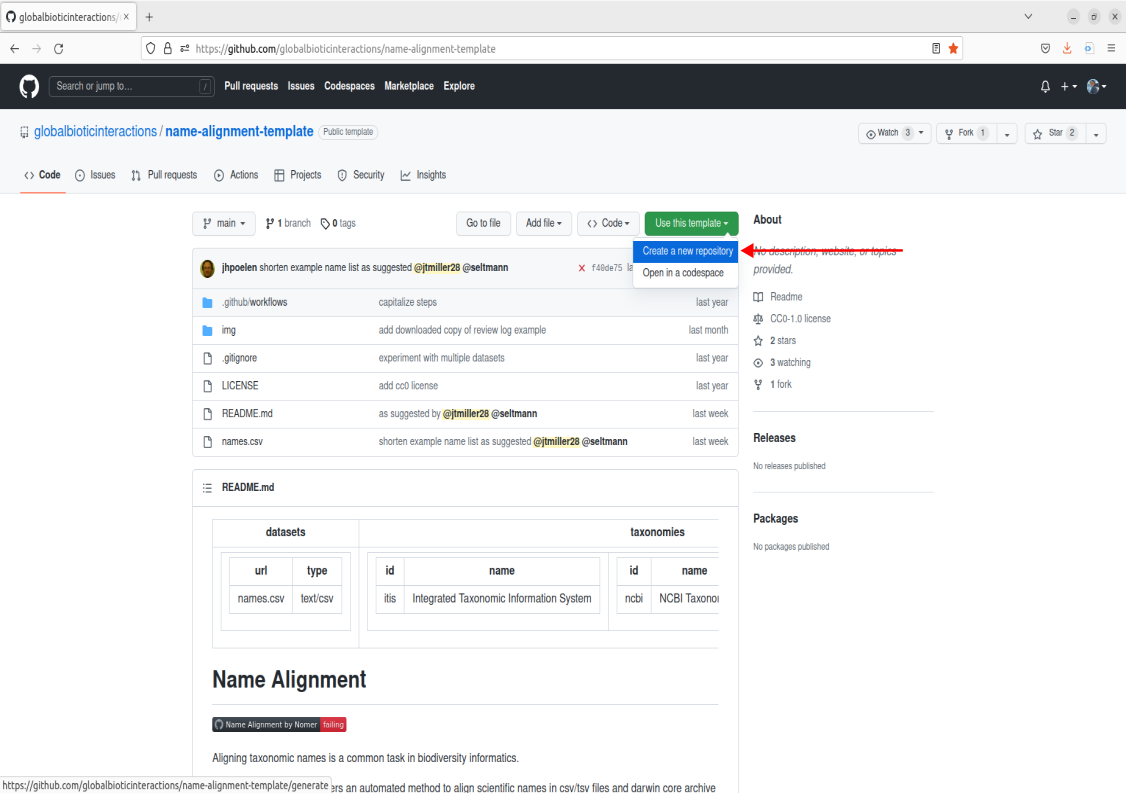
Navigate over to the search bar in the top left corner. Enter in the following:
globalbioticinteractions/name-alignment-templateClick on the option that says “jump to”. This will bring you to the landing page for the name-alignment-template. This is the official repo that holds the tool made by Jorrit. Now we want to create a personal copy of the tool by creating a new repository based upon this template. To do this, navigate over to the green button in the middle right of the screen that says “Use this template”, click it, and now you should see an option to create a new repository. Go ahead and click that.
From here, you will be prompted to fill out some information for creating a new repository from the name-alignment-template. You will want to create some type of repo name, give it a description, choose the privacy setting you prefer, and then click the green button “Create repository from template”.
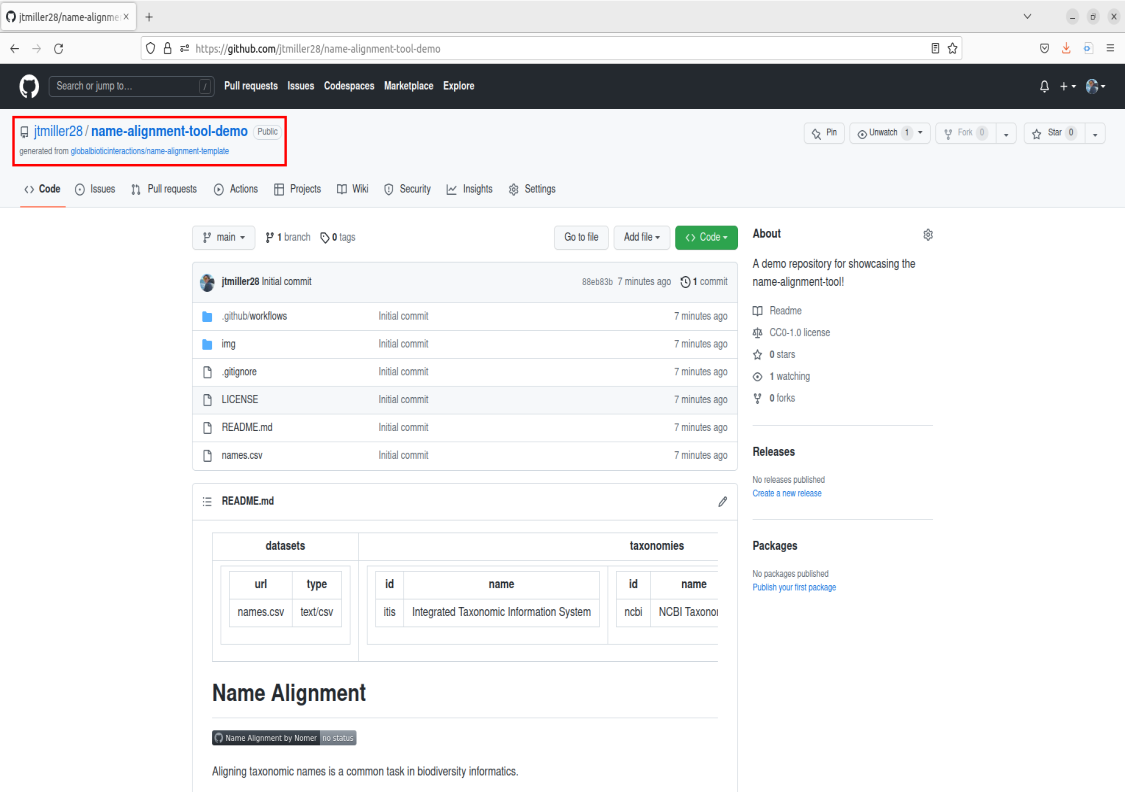
Congrats! Now you should have your very own copy of the name-alignment-template.
Navitgating my New Repo
Now lets cover some basics for guiding ourselves around the repository. There is a lot of information on this webpage, but for our purposes we are going only highlight what is necessary to operate the tool. First, you see the directory of where your repository is stored by looking in the top left corner. It should be structured as username/name-of-repository. Take a look at this and make sure it is structured like this to confirm you made a copy of the template!
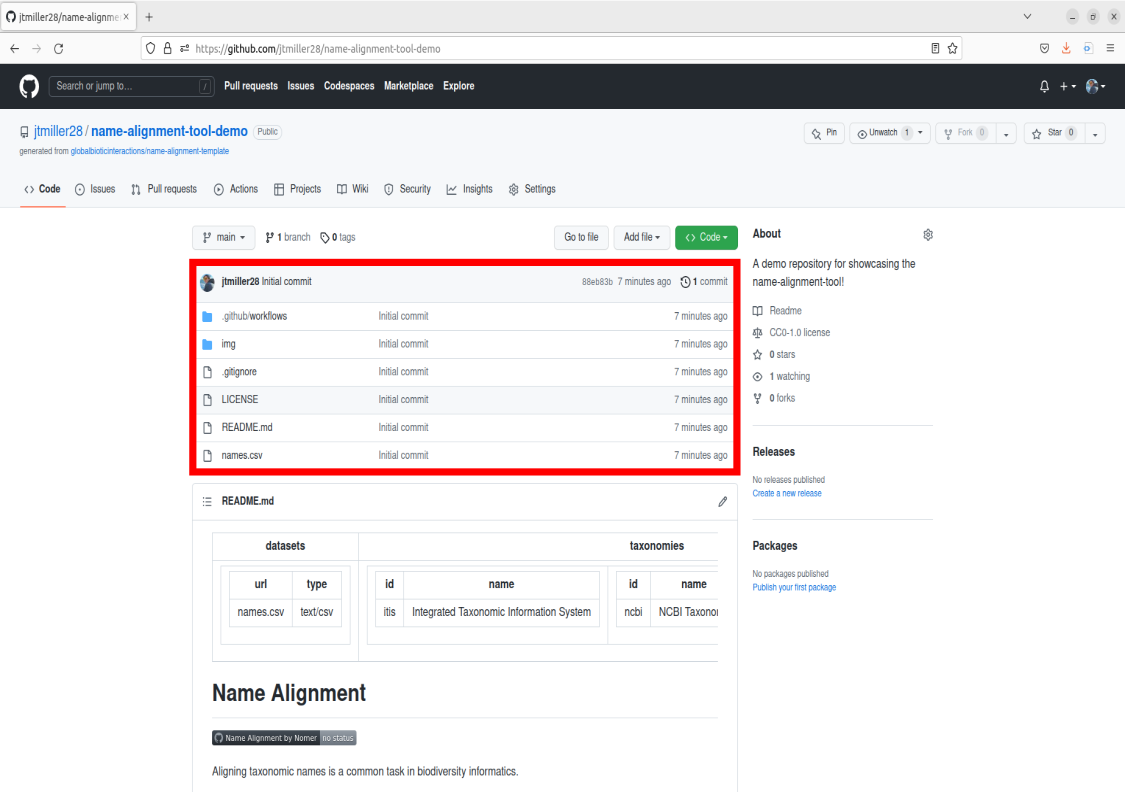
Recall that a repository is basically a folder containing files. The files can be visually seen on your repo’s main Code page in the center box. This file box contains other info such as sub-folders/directories in the repositiory as well as time stamps for when they were last modified.
If you scroll down you should see a section with a subheader called README.md. Readmes are typically designed as front pages for repositories to display helpful information about how to use the repository. Typically readmes are designed to just convey information, however today we’ll be exploring a unique application of editing the readme in order to use the name-alignment-tool, but more on that later! For the purposes of this example tool, we can also think of the README as the description of what our Name-alignment-tool does.
Continue on to the next lesson for a showcase of resolving names using your new tool!
Key Points
Github is an important tool for collaboration
Accessing the tool and using it through githubs interface can be quite easy provided you can navigate the basics!
Using the name-alignment-tool
Overview
Teaching: 15 min
Exercises: 10 minQuestions
How do I interact with this tool?
How can I change the names that the tool resolves?
How can I choose which catalogues to align names to? What are my catalogue options?
How can I access my output? What do these outputs mean?
Objectives
Understand how to navigate the tool
Learn the process of adding files containing names for matching, requirements, and accessing the tool’s output.
Setup - remote directory & file structure
For the tool to interact and read our files, we need either a file stored remotely on this github repository or a dwc archive link. For todays demonstration lets look at how we can upload a list of names to our remote directory for the github repository
Note: “Remote Directories” You can think of this as a google drive, basically a file storage system that operates on servers or the “cloud”. Like google drive, you can create folders, add and organize files.
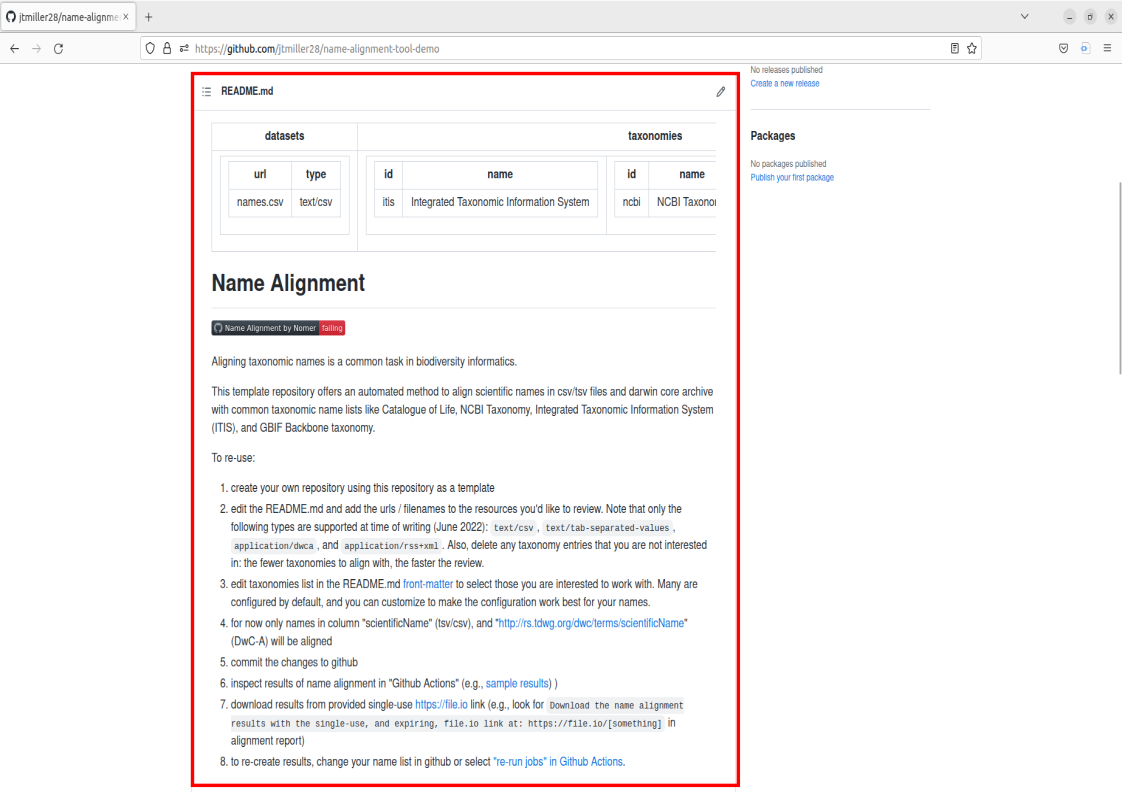
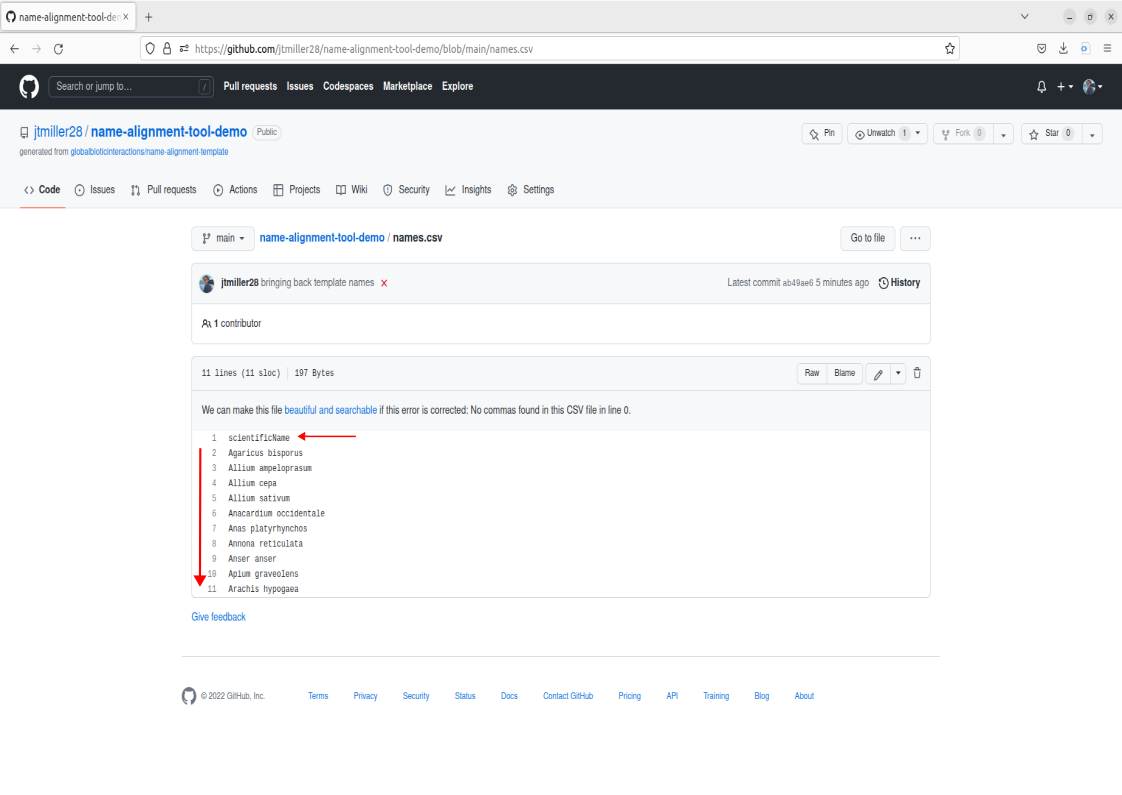
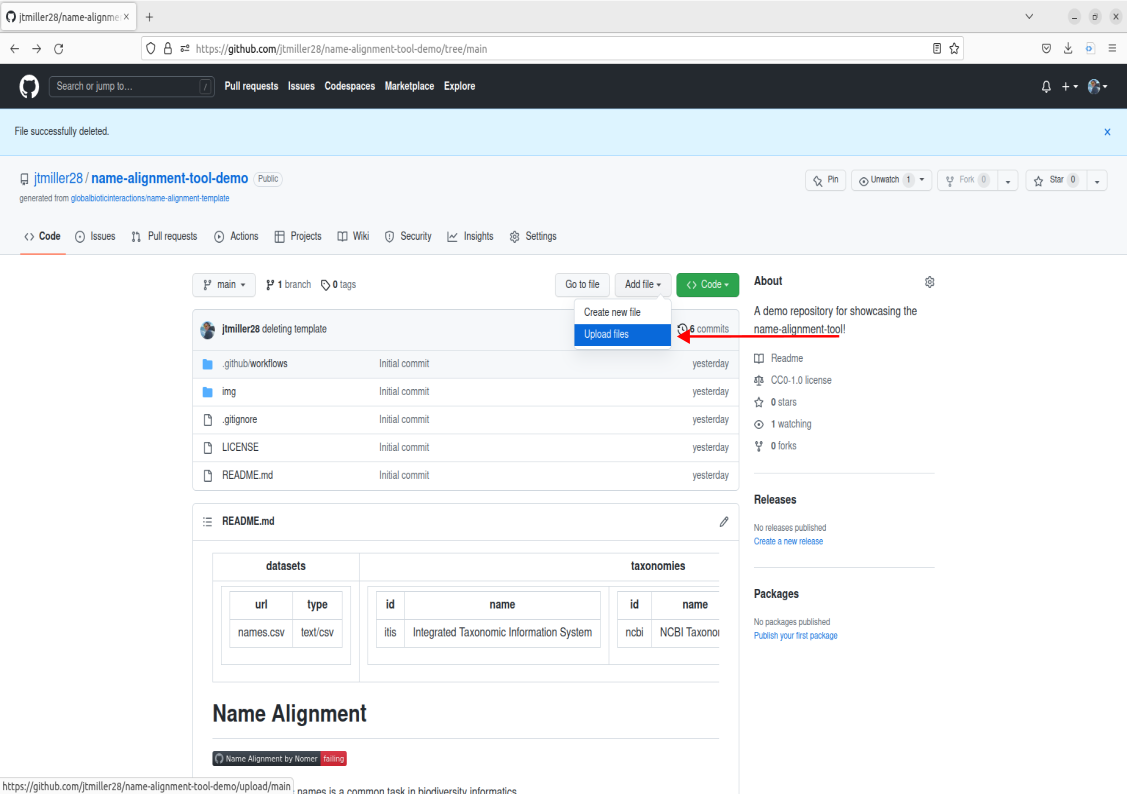
Lets first take a look at the file in our remote repository called names.csv. Navigate to the files and go ahead and click on it.
This is an example of how the names should be formatted when inputted into the tools. The two main things to notice that are a requirement for your names to align correctly are:
- the “scientificName” field must be present in the first row.
- from the second row on, each name will occupy its own row
Note that scientificName must be present, but it can be in any column of the dataset. The tool will select only the column labelled scientificName.
This example names.csv file contains names we could match, but lets try uploading a different list of names containing bees. We can get rid of the old name file by either deleting it, to do this click on the trashcan icon in the right corner of the file.
Example names
example file: names.csv
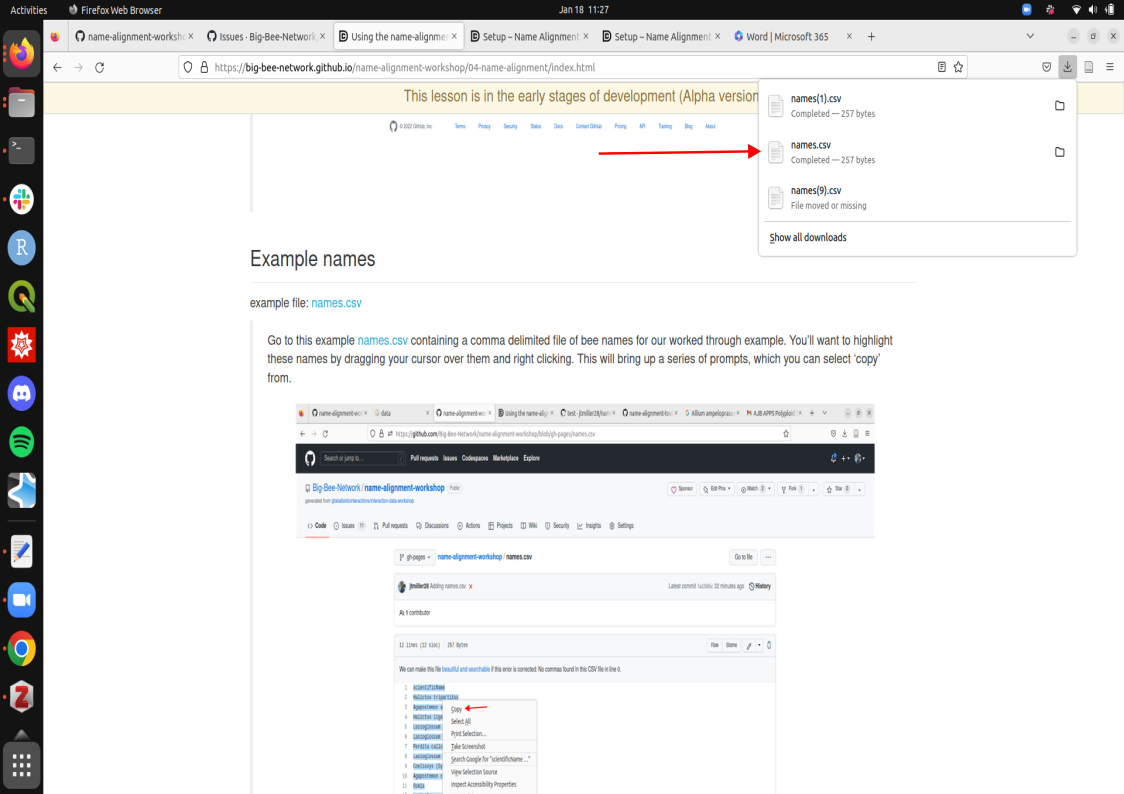
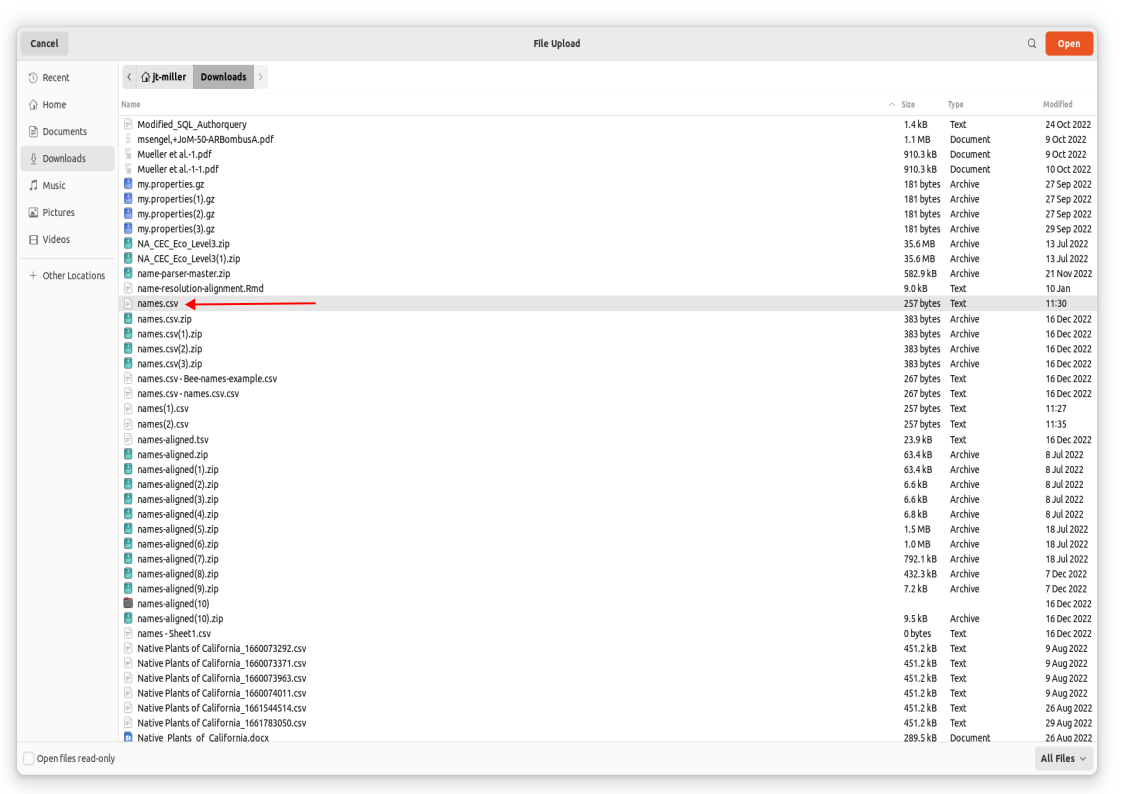
Click on this link names.csv containing a comma delimited file of bee names for our worked through example. This will download a list of bee names to your personal device.
This file is now downloaded onto your local machine. We now would like to upload these names onto github’s remote directory (where that file box sits).
this is done by:
- Click Add File, then select ‘Create new file’.
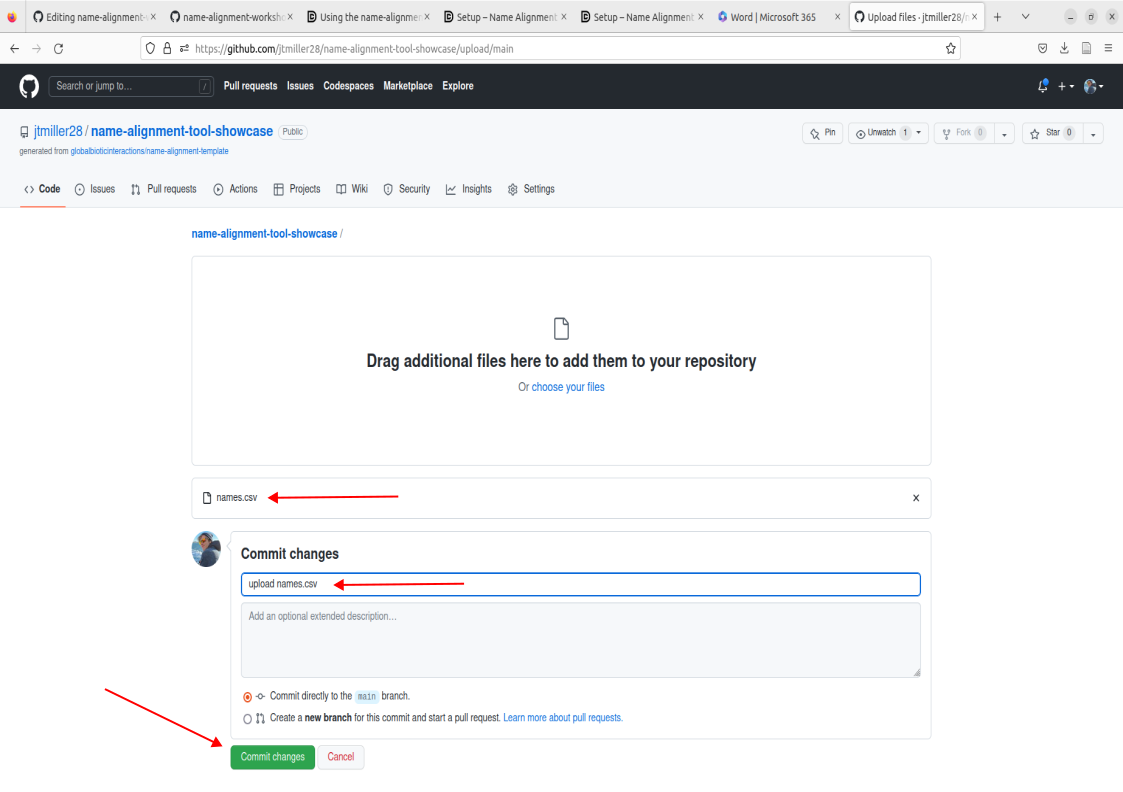
- This will take you to a commit screen where we can select that file on our local machine. Click on “choose your files”. You should be able to find your names.csv in your Downloads folder.
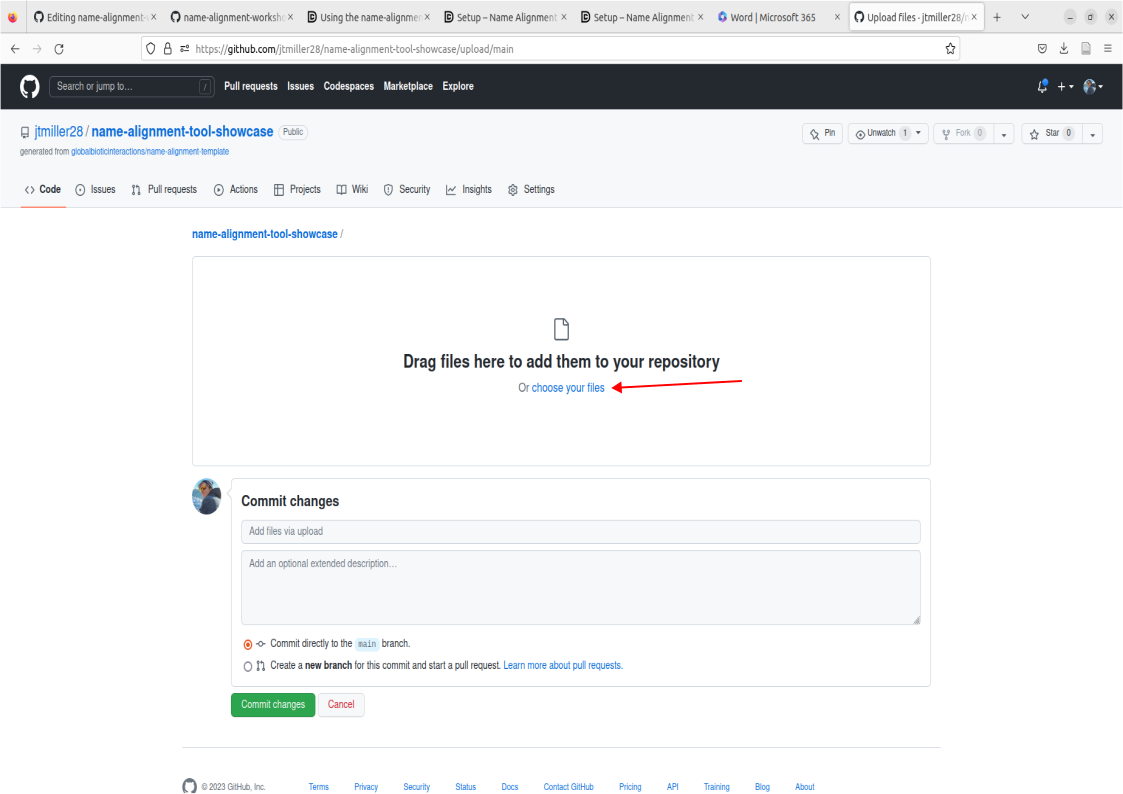
- Important! Make sure the file is named “names.csv”, this is a requirement for the purposes of this tutorial. Now proceed to commit these changes after writing a commit message of your choosing.
- Now we you should have names.csv located within the file box on your repo, go ahead and check to see if its there.
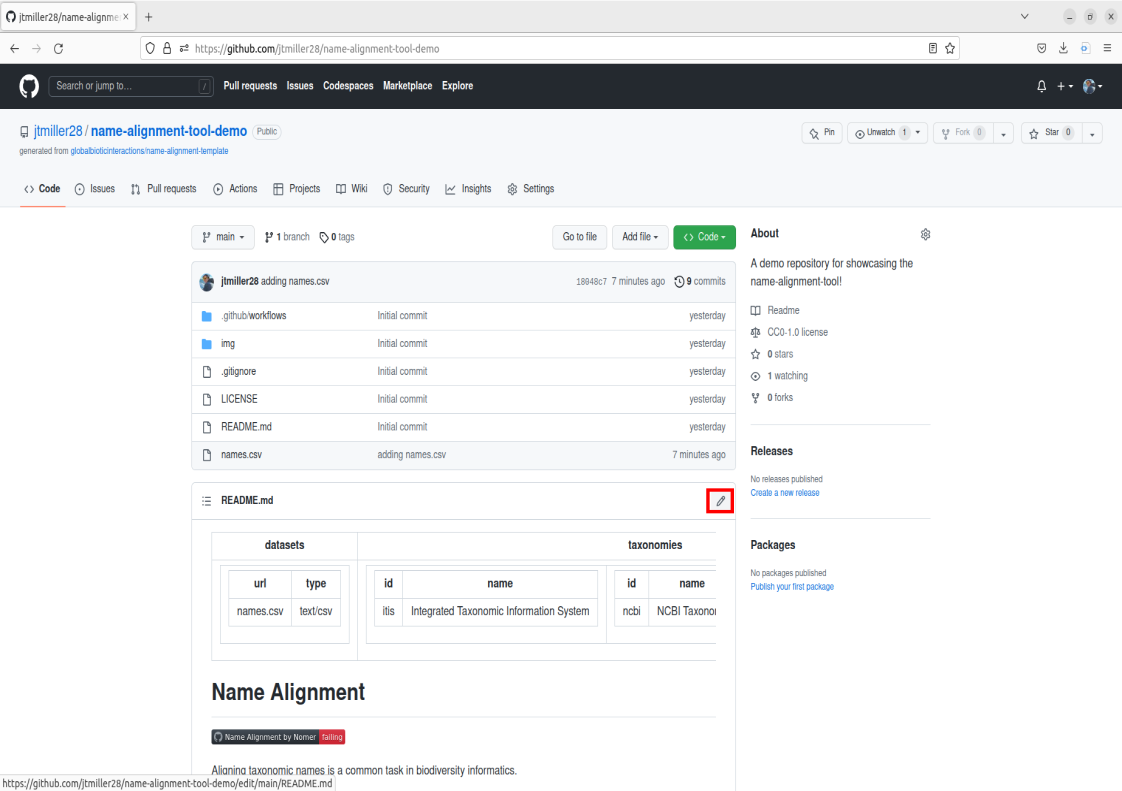
You should now see the file in the Repo called names.csv! This means the file is now stored remotely on this github repo.
Running name alignment
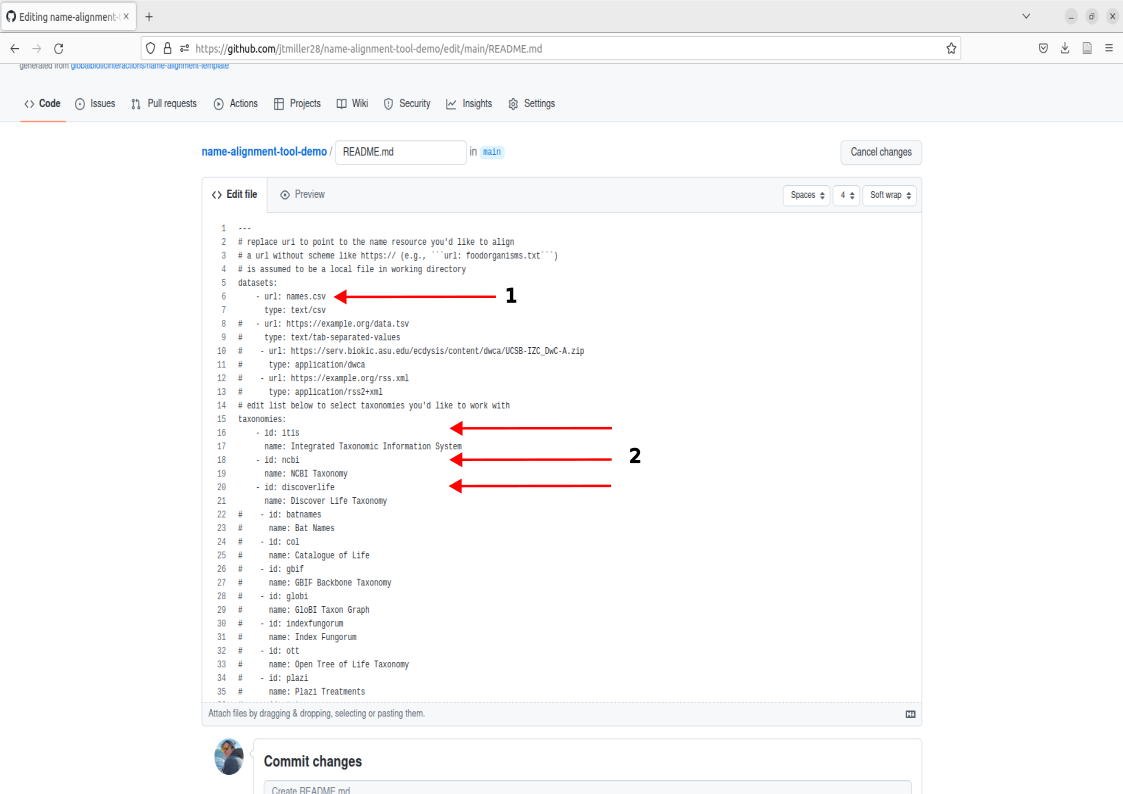
To use the tool we will be commiting changes to the repository’s description (ReadMe). To do this we’re going to select the pencil icon located on the top right of the description box.
This will bring you to a new page where we can edit the description. Some things to take note of are:
- The subsection called datasets: Here the tool will search the repository’s directory for the name of the file we put here. names.csv is already placed so no edits will be needed here.
- The subsection called taxonomies: Here, you can edit which catalogues are used for matching by either adding or removing the ‘#’. For example, if I didn’t want to use the catalogue itis I could add ‘#’ in front of the id and name field for that catalogue. For the purposes of this worked example we already have the three catalogues, itis, ncbi, and discoverlife active and ready to match.
You can now run name alignment. Note that you technically cannot commit/save if there are no changes to the description, so just add a ‘#’ to the end of the document since we know that won’t do anything. Write a commit message like “Run name-alignment” and Commit changes.
Alignment will be processing in the background of the github, however we can also see the progress happening in real time by looking at our most recent commit history. To do this we can select the button called ‘Name Alignment by Nomer’
This will bring you to a page indicating that the alignment is operating by your commit history. The circle icon next to the commit message will either be yellow indicating alignment is in progress or green indicating alignment has finished. You can click on this commit message to access the alignment.
While in progress, you can view the alignment tool in action by clicking on the “align” button.
This will bring you to the background process where the name-alignment-tool is working on aligning names.
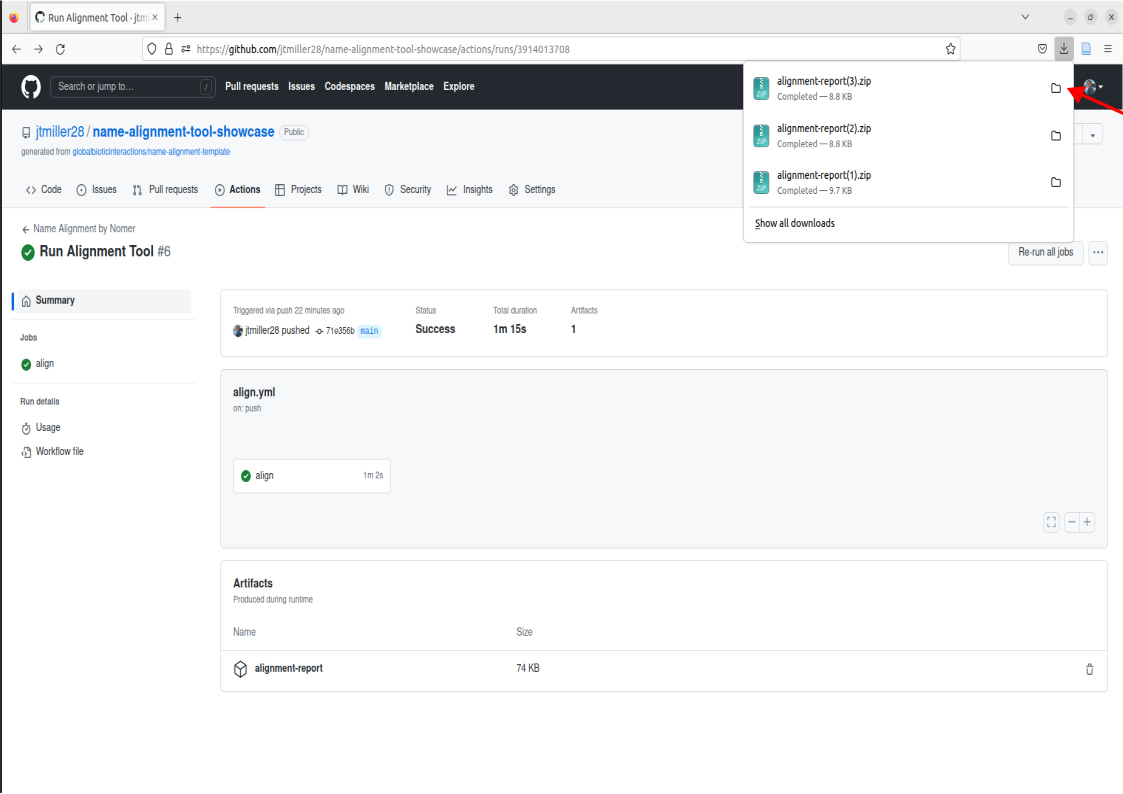
Once alignment has finished, we can now access the alignment report by clicking on the tab called “alignment report”
This will download the names as a zip file to your computer’s downloads. You can then access the alignment by clicking on the downloaded file.
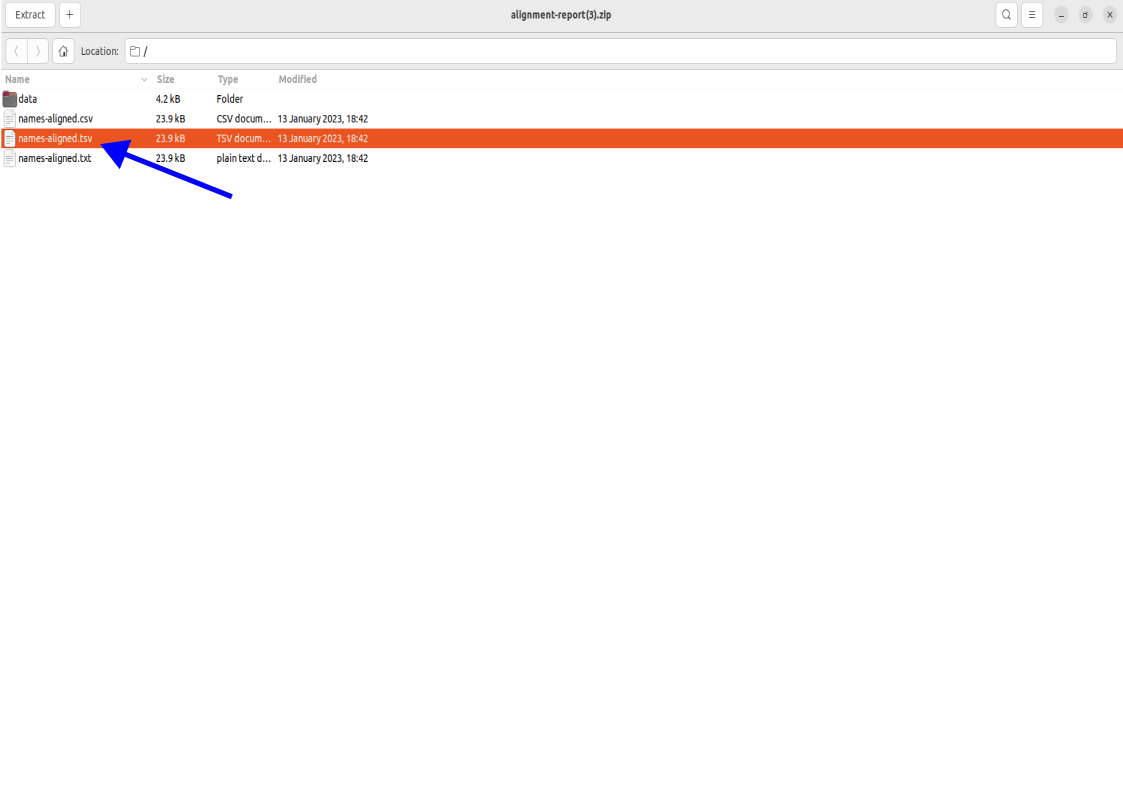
Accessing Aligned Names
To open the names, click on the names-aligned.tsv
Up next we’re going to interpret our aligned names
Key Points
Knowing the basic steps of adding data, data requirements, changing the readme, and accessing the outputs will give you full access to the tool’s aligning capabilities.
What do these resolutions actually mean?
Overview
Teaching: 10 min
Exercises: 10 minQuestions
How should I read the output given by the name-alignment-tool?
Which is the ‘correct’ name alignment?
What if a name isn’t matching to any of the catalogues?
Objectives
Understand how to interpret the name-alignment output
Understand the basics in how names are aligned, and how this can impede resolution
Interpreting the output
Before showing the downloaded output: Let’s think a bit about what is happening under the hood of name alignment. Below is a diagram denoting the steps taken to align our names by the tool. Here, the tool takes in the name list, processes and aligns each name against the catalogues of choice. The tool then formats and returns a table of our initial names and their aligned relations.
Below is a sample table output of name alignment with some highlighted fields. Please note that the name-alignment tool is constantly recieving updates based upon feedback, so there likely will be more fields in your own outputs.
There is quite a bit here so lets break down the fields that are of particular interest:
- The field providedName represents the scientificName field we provided in our input to the tool.
- The field parsedName represents the scientificName after undergoing parsing, parsing is a process where extranoeous information is truncated out of the name if a matching scheme supports it (e.g. Authorship/Authority). Please note that subgenuses are parsed out change?
- The field alignRelation can have three possible values:
- HAS_ACCEPTED_NAME - a name is matched and is up to date/current
- SYNONYM_OF - name is matched, but has been identified as synonymous to another name
- NONE - indicates that there was no match found in that particular catalogue.
- the field alignedExternalId indicates the catalogue the name was matched to. These are often abbrievated containing the ID or url within the catalogue to reference that name alignment.
- the field alignedName is the name relation matched to the parsedName.
- the field alignedRank is the taxonomic level of the alignedName (i.e. Genus, species, etc).
- the field alignedPath shows a ‘piped’ list of the alignedNames taxonomic designation. ` Domain | Kingdom | Phylum | Class | Order | Family | Genus | Species`
Conclusion & Further Discussion
That about wraps up the basics you’ll need to use this tool. We hope that you found this tutorial helpful and that you can use this tool to ease your workflow in keeping the collections up to date. If you are interested in more details of how names are aligned be sure to visit Nomer github repo, and if you have any questions/problems with the tool you can raise an issue on the name-alignment-tool repo.
When writing an issue, its best to be explicit about a the problem you run into or are trying to ask. Vague problems are much harder to solve after all! Here’s an example of an issue:
Things to note:
- Write a brief but concise description of the problem you are running into
- (Not included in this example, but better practice) A brief description on what you think should be intended. This is also a great kickstarter that can lead into some really interesting discussions!
- Copy-Paste either outputs or visuals highlighting the issue (in this example I included the name-alignment-tool table outputs).
Discussion
Thoughts about the tool or name alignment in general?
In case we have extra time!
Now that we’ve learned the basics of the tool, lets go ahead and try some names! Feel free to post any scientificName in the chat, and lets see how name alignment handles it! Complicated ones (ones known to have unstable taxonomy are encouraged!) Here are some example ones
| scientificName |
|---|
| Acanthalictus dybowskii |
| Adanthidium discophorum |
| Agapostemon azarae |
| Dufourea versatilis rubriventris |
| Eoanthidium (Eoanthidiellum) elongatum |
Key Points
Resolution is subjective to the catalogue, therefore the aligned names should be treated as hypotheses rather than true standards
Name matching with the name-alignment-tool is accurate yet naive, the tool won’t attempt to interpret mistakes
Data Sources: Taxonomic Name Review
Overview
Teaching: 10 min
Exercises: 10 minQuestions
How does GloBI interpret taxonomic names?
How can I find taxonomic names that GloBI didn’t understand?
Does GloBI use single taxonomic backbone?
Objectives
Understand how GloBI links provided names to existing taxonomic naming schemes
Know to where to go to find, and discuss, suspicious taxonomic names
Getting started
At the end of this time we will regroup and report back the other workshop participants about what we did in this breakout group. Who would like to be the person/s who report back for the breakout group?
Let’s collaboratively take notes in the Google Document. The link to the document is in the chat.
Goal
The goal of this lesson is to understand how, and why, GloBI indexes and links taxonomic names.
Why Link Taxonomic Names?
Taxonomic names are used in literature and datasets to classify living organisms. To increase the discoverability of species interaction data, GloBI links and indexes provided taxonomic names to enable queries like: which mammals (Mammalia) are known the host ticks (ticks)? Also, taxonomic name linking enables retrieval of key images and common names to help provide a context and make it easier to interpret an interaction claim.
Taxonomic Name Linking Challenges
However, significant challenges exist to interpret and link taxonomic names, especially when dealing with datasets from different eras, authors, and disciplines.
These taxonomic name interpretation challenges include, but are not limited to:
- Use of common names instead of scientific names (e.g., “kip” (Dutch) or “chicken” (English) vs. “Gallus gallus domesticus”)
- Typos (e.g., “Gallvs gallus” instead of “Gallus gallus”)
- Ambiguous names (e.g., “Anura” is a genus of flowering plants in the daisy family of as well as an order for frogs).
- Outdates/ disputed names (e.g., taxonomic revisions re-interpret classifications and re-assign names)
- Incomplete hierarchies (e.g., data sources provide species name, but no higher order taxonomic ranks)
Taxonomic Name Links
GloBI uses existing taxonomic name parsing and resolving tools to help find reasonable links between provided (or verbatim) names from data sources and existing taxonomic name lists. Rather than using a single taxonomic backbone, a variety of name sources is used. These name sources include, but are not limited to: Integrated Taxonomic Information System (ITIS),
![]() World Register of Marine Species (WoRMS),
World Register of Marine Species (WoRMS), NCBI Taxonomy,
Wikidata Taxonomy,
Encyclopedia of Life (EOL) species pages,
FishBase,
SeaLifeBase,
iNaturalist Taxonomy,
![]() GBIF Backbone Taxonomy, and
GBIF Backbone Taxonomy, and ![]() Plazi’s TreatmentBank.
Plazi’s TreatmentBank.
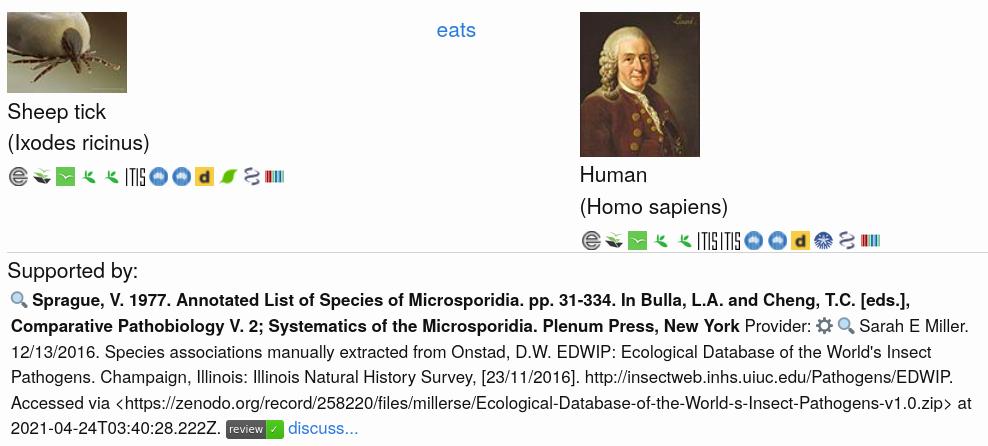
Exercise 1. Discover Taxonomic LinksNote the icons representing the various taxonomic name schemes in the screenshot of an interaction claim of a sheep tick (Ixodes ricinus) eating a human (Homo sapiens).
Click on the image to try and reproduce the results. Then, discover the taxonomic linkages using the icons below the image. Note what project provides the images for the taxa. Also, record some examples of the urls pointing to the taxonomic name resources by right-clicking on the icon and copy-pasting link using “copy link location” (or similar). What do you notice?
GloBI’s Taxonomic Name Linking Process
The process to index and resolve taxonomic names currently consists of two phases.
Phase 1. Create a Taxonomic Name Map (aka GloBI’s Taxon Graph)
a. extract taxonomic names/ids from data sources
b. pre-process and parse names/ids
c. match names/ids against existing names resolvers/name lists
d. version and publish (updated) taxon name map/graph
Phase 2. Create Search Index
a. load specific version of taxon name map/graph
b. on encountering mapped taxonomic names/id, add to extra information index
c. on countering unmapped name/id, tag name with “no:match”
d. version and publish interaction search index
Want to Learn More about GloBI's Taxonomic Name Matching?Visit https://globalbioticinteractions.org/process to learn more about taxonomic name matching. Also, you might want to have a look at a recent publication of GloBI’s Taxon Graph at https://doi.org/10.5281/zenodo.755513 .
Finding Suspicious or New Names
If GloBI encountered a name that has not yet been successfully mapped in GloBI’s Taxon Graph, the name is labeled with “no:match” in the search engine. Now, we can use this label to find interaction records that include names that are new to GloBI or failed to match to supported taxonomic name schemes.
So, “no:match” names might include names that contain typos, but may also include names that are valid, but have not yet been included in GloBI’s Taxon Graph.
Exercise 2. Finding Suspicious NamesShow three ways you can find suspicious names using GloBI tools.
Feedback Loops
Exercise 3. Reporting a suspicious name or starting a discussionAdd add examples on how to reach out to peers to discuss suspicious names or records.
Key Points
Taxonomic name linking facilitates discovery, review, and interpretation, of interaction records
GloBI uses a versioned taxonomic name map to map verbatim names into known taxonomic schemes
GloBI attempts to provide reasonable links using a controlled and iterative process
GloBI taxonomic name linking process is likely imperfect and subjective
Data Sources: Interaction Data Record Review
Overview
Teaching: 15 min
Exercises: 5 minQuestions
How can I understand how GloBI interpreted my dataset?
Can GloBI help me improve my dataset?
Objectives
Find out where GloBI reviews are located.
Find out how to explore GloBI reviews
Goals
The goal of this lesson is to introduce you to data reviews in GloBI.
Getting started
At the end of this time we will regroup and report back the other workshop participants about what we did in this breakout group. Who would like to be the person/s who report back for the breakout group?
Let’s collaboratively take notes in the Google Document. The link to the document is in the chat.
What is a review
A review in GloBI is an output that lets us know how GloBI interpreted or viewed the data being indexed. It is an opportunity to see if you agree with the interpretation or find issues in the data that can be corrected. It also provides some cool statistics about the number of interaction records indexed from a particular dataset. Reviews are done by dataset only. Reviews are useful for people who are submitting data, data curators, or anyone who wants to know more about a particular dataset.
Finding the reviews
Remember we explored in working with data sources that all GloBI data sources are listed on the sources page. In addition, providers for Terrestrial Parasite Tracker have their own webpage.
Let’s examine a GloBI data review. Visit https://globalbioticinteractions.org/sources and locate the ucsb-izc collection. Click on the
badge.
What is what? An overview of the data.
_____ _ ____ _____ _____ _
/ ____| | | _ \_ _| | __ \ (_)
| | __| | ___ | |_) || | | |__) |_____ ___ _____ __
| | |_ | |/ _ \| _ < | | | _ // _ \ \ / / |/ _ \ \ /\ / /
| |__| | | (_) | |_) || |_ | | \ \ __/\ V /| | __/\ V V /
\_____|_|\___/|____/_____| |_| \_\___| \_/ |_|\___| \_/\_/
| | | ____| | |
| |__ _ _ | |__ | | |_ ___ _ __
| '_ \| | | | | __| | | __/ _ \| '_ \
| |_) | |_| | | |____| | || (_) | | | |
|_.__/ \__, | |______|_|\__\___/|_| |_|
__/ |
|___/
Review of [globalbioticinteractions/ucsb-izc] started at [2021-04-26T06:04:32+02:00].
Review of [globalbioticinteractions/ucsb-izc] included:
- 1440 interaction(s)
- 25 note(s)
- 1442 info(s)
[globalbioticinteractions/ucsb-izc] has 25 reviewer note(s):
7 found unsupported interaction type with name: [Visiting]
6 source taxon name missing: using institutionCode/collectionCode/collectionId/catalogNumber/occurrenceId as placeholder
3 found unsupported interaction type with name: [Sitting on]
3 found unsupported interaction type with name: [Hovering over]
2 found unsupported interaction type with name: [Feeding on]
1 found unsupported interaction type with name: [Visitng]
1 found unsupported interaction type with name: [visiting]
1 found unsupported interaction type with name: [Tended by]
1 found unsupported interaction type with name: [Next to]
1440 interaction(s) is number of interactions indexed by GloBI from this dataset.
25 note(s) or rows in the dataset that are flagged and might be interesting to have a look at.
1442 info(s) or information about biotic interaction indexing process. These are not flagged records, but more like comments or data logging.
6 source taxon name missing where the taxon name field is blank or empty
unsupported interaction type indicates that no mapping is defined by the data source. Terms are defined by linking to the Relations Ontology.
What is what? I want more information.
GloBI data reviews are packaged in downloadable text files (tab and comma delimited). The review-sample files are small enough to quickly view.
This review generated the following resources:
- review.svg (review badge) https://depot.globalbioticinteractions.org/reviews/globalbioticinteractions/ucsb-izc/review.svg
- review.tsv.gz (data review) https://depot.globalbioticinteractions.org/reviews/globalbioticinteractions/ucsb-izc/review.tsv.gz
- review-sample.tsv (data review sample tab-separated) https://depot.globalbioticinteractions.org/reviews/globalbioticinteractions/ucsb-izc/review-sample.tsv
- review-sample.json (data review sample json) https://depot.globalbioticinteractions.org/reviews/globalbioticinteractions/ucsb-izc/review-sample.json
- review-sample.csv (data review sample csv) https://depot.globalbioticinteractions.org/reviews/globalbioticinteractions/ucsb-izc/review-sample.csv
- indexed-interactions.tsv.gz (indexed interactions) https://depot.globalbioticinteractions.org/reviews/globalbioticinteractions/ucsb-izc/indexed-interactions.tsv.gz
- indexed-interactions.csv.gz (indexed interactions) https://depot.globalbioticinteractions.org/reviews/globalbioticinteractions/ucsb-izc/indexed-interactions.csv.gz
- indexed-interactions-sample.tsv (indexed interactions sample) https://depot.globalbioticinteractions.org/reviews/globalbioticinteractions/ucsb-izc/indexed-interactions-sample.tsv
- indexed-interactions-sample.csv (indexed interactions sample) https://depot.globalbioticinteractions.org/reviews/globalbioticinteractions/ucsb-izc/indexed-interactions-sample.csv
- indexed-names.tsv.gz (indexed names) https://depot.globalbioticinteractions.org/reviews/globalbioticinteractions/ucsb-izc/indexed-names.tsv.gz
- indexed-names.csv.gz (indexed names) https://depot.globalbioticinteractions.org/reviews/globalbioticinteractions/ucsb-izc/indexed-names.csv.gz
- indexed-names-sample.tsv (indexed names sample) https://depot.globalbioticinteractions.org/reviews/globalbioticinteractions/ucsb-izc/indexed-names-sample.tsv
- indexed-names-sample.csv (indexed names sample) https://depot.globalbioticinteractions.org/reviews/globalbioticinteractions/ucsb-izc/indexed-names-sample.csv
- indexed-citations.tsv.gz (indexed citations) https://depot.globalbioticinteractions.org/reviews/globalbioticinteractions/ucsb-izc/indexed-citations.tsv.gz
- indexed-citations.csv.gz (indexed citations) https://depot.globalbioticinteractions.org/reviews/globalbioticinteractions/ucsb-izc/indexed-citations.csv.gz
- nanopub.ttl.gz (interactions nanopubs) https://depot.globalbioticinteractions.org/reviews/globalbioticinteractions/ucsb-izc/nanopub.ttl.gz
- nanopub-sample.ttl (interactions nanopub sample) https://depot.globalbioticinteractions.org/reviews/globalbioticinteractions/ucsb-izc/nanopub-sample.ttl
- review.zip (review archive) https://depot.globalbioticinteractions.org/reviews/globalbioticinteractions/ucsb-izc/review.zip
Get a glimpse of the review in Google Sheets
1 - Import a review-sample file into Google Sheets
2 - Download the full review archive 3 - Import the indexed-interactions-sample.tsv into Google Sheets
Exercise 1: Find Data SourcesVisit https://globalbioticinteractions.org/sources and locate the USNM Ixodes Collection, Seltmann’s > Tick Interaction Database, or iNaturalist observation records. For each, click on the
badge to open the review page. For one of the datasets: 1 - find out how many interactions have been indexed for each dataset. 2 - import the indexed-interactions-sample.tsv into Google Sheets
Key Points
There are many ways to access GloBI reviews.
GloBI reviews can help data managers better understand their data and how GloBI interprets it.